Segmentation#
The image segmentation module allows researchers to quickly identify cells or nuclei by selecting pre-trained segmentation models. Users can choose a model from the available options and apply it to images that are opened and selected.
Cellpose: Remote Computation
All of the models run locally on your own device EXCEPT the Cellpose model. This has to do with the difficulty converting the Cellpose model, which utilized many custom functions, to a usable TFJS model. To get around this, we connect to the BioEngine, maintained by the AICell Lab, server to run this model. It is possible that the server may stop responding and you will get an error during inference. If this happens you may just need to wait a little while for the server to reboot and try again.
Overview#

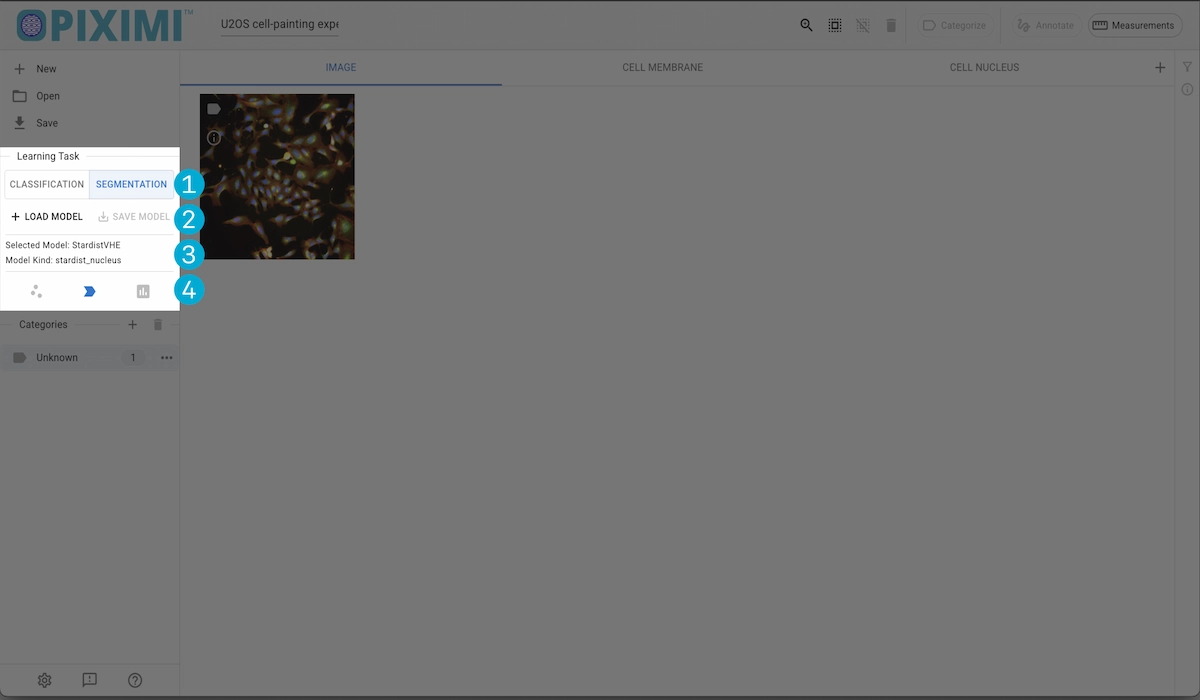
Task Selection
Model I/O
Model Description
Model Operations
Task Selection#
Switch between classification and segmentation tasks. The tasks will operate on the currently displayed Kind.
Model I/O#
Model Selection#
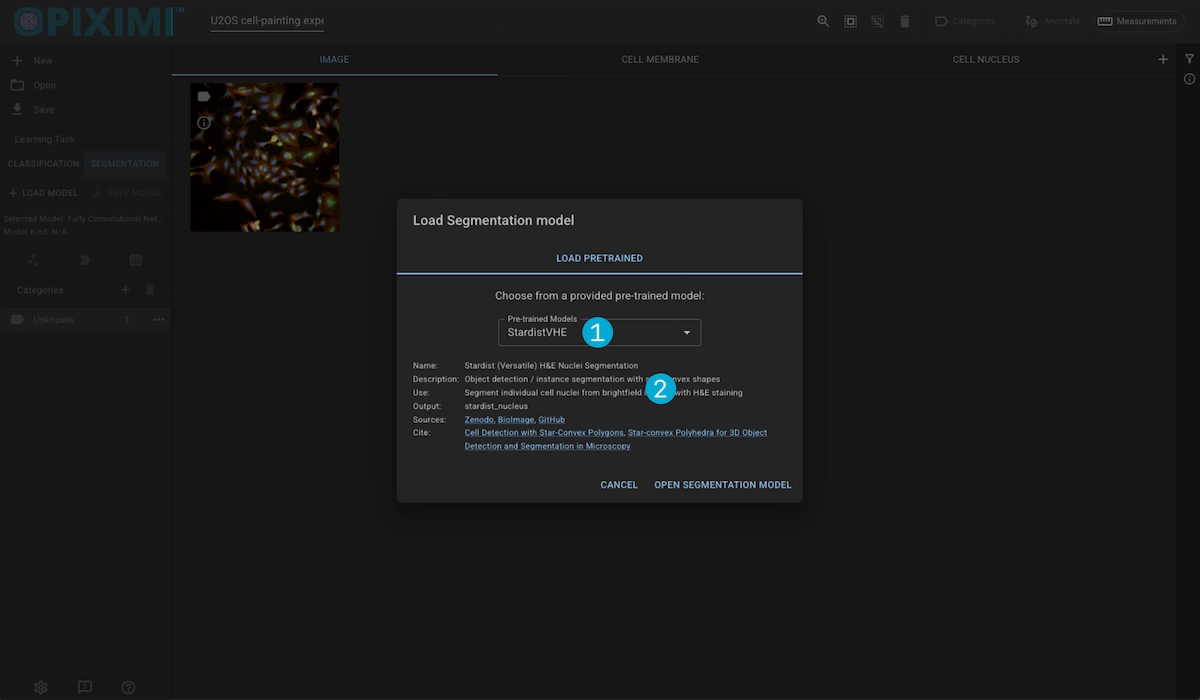
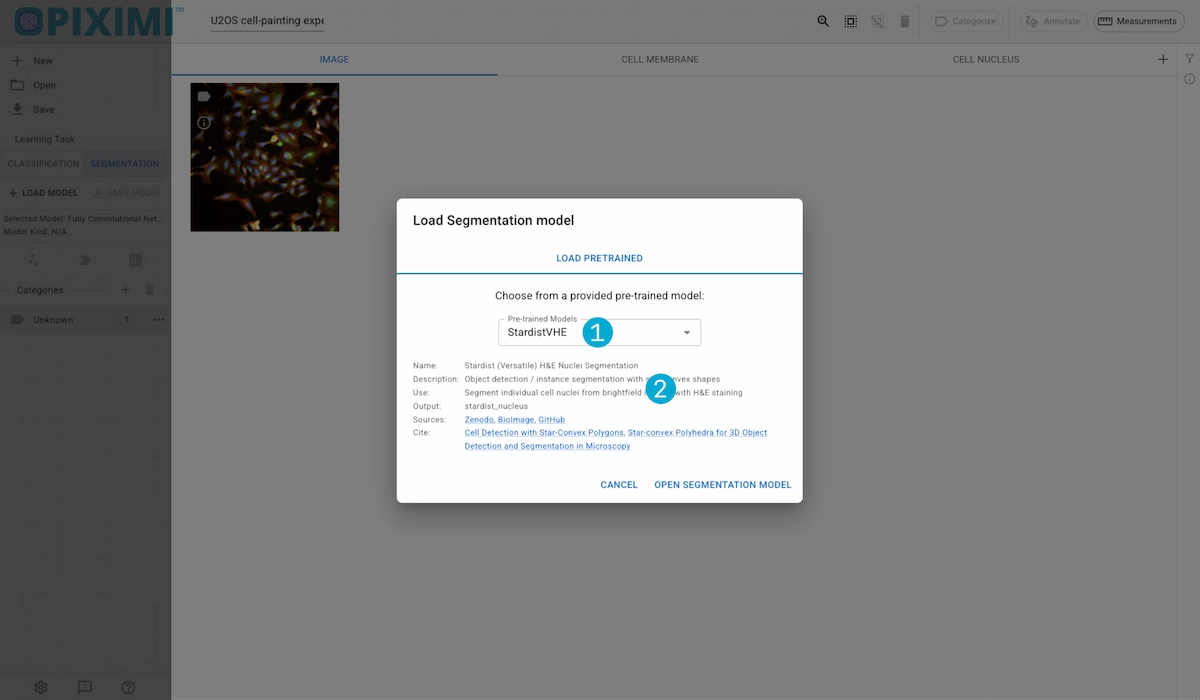
1. Model Selection
Piximi provides several segmentation models to choose from:
Cellpose (remote)
Stardist (fluo, VHE)
Gland Segmentation
COCO-SSD
More informatino about the models can be found in the Segmentation Tutorial.
2. Model Details
Displays information about the model, its sources, the Kind output, and a potential use-case.
Model Description#
Display the name of the currently selected model, as well as the name of the Kind of object it identifies.
Model Operations#
Fit#
We are currently working on support for training a segmentation model.
Predict#
Clicking the Predict button will begin running inference on the images/object of the displayed Kind, using the selected model. A new Kind will be created and populated with the identified objects.
Evaluate#
Since we do not support segmentation model training at this time, there is nothing to evaluate.
See our Segmentation Tutorial for usage information.