Object Classification
Contents
Object Classification#
Currently, Piximi can only classify entire images. We are working on adding deep learning based segmentation that would enable users to classify individual cells or other segmented regions of interest within an image. This approach will be useful for recording phenotypes of cell subpopulations within a larger, heterogenous population, for example.
While we work on adding segmentation, it’s currently possible to use CellProfiler to segment individual cells and create single cell cropped images. These images of individual cells can be then be opened in Piximi and then categorized.
The following example will demonstrate how the image crops for the Piximi U2OS Cell Crops example project were made using CellProfiler. The images we will use to recreate this dataset are from the BBBC013 cytoplasm-nucleus translocation dataset from the Broad Bioimage Benchmark Collection. Within this dataset, a cytoplasmic GFP signal is considered as positive and a nuclear GFP signal as negative. The CellProfiler pipeline used in the following example can be downloaded here for CellProfiler version 4.2.2. For CellProfiler 4.2.1 or earlier, download this pipeline. Alternatively, follow this guide to learn how to create this pipeline from scratch.
Create cell crops with CellProfiler#
For this example, we will create a CellProfiler pipeline that will first identify nuclei using the IdentifyPrimaryObjects module, followed by cell identification with the IdentifySecondaryObjects module. Following this, we will then create a multichannel RGB image with DNA colored blue and GFP colored green. Finally, these mulltichannel RGB images will be cropped based on the previously identified cell object using the SaveCroppedObjects module.
1. Import images#
To begin, drag and drop your images into the Images input module of CellProfiler.
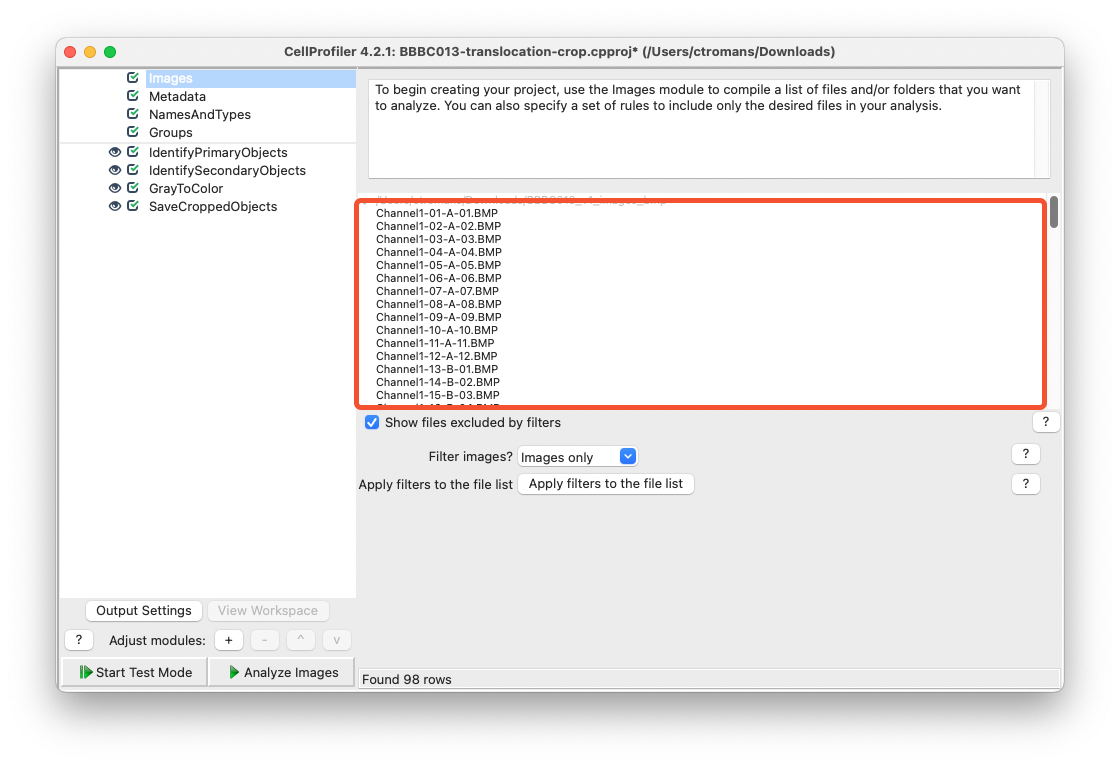
Fig. 19 Drag and drop your images into the Images input module.#
Next, select appropriate rules to categorize your files in the NamesAndTypes input module. For images that contain Channel2 in their filename, assign the name rawDNA. For images that contain Channel1 in their filename, assign the name rawGFP.
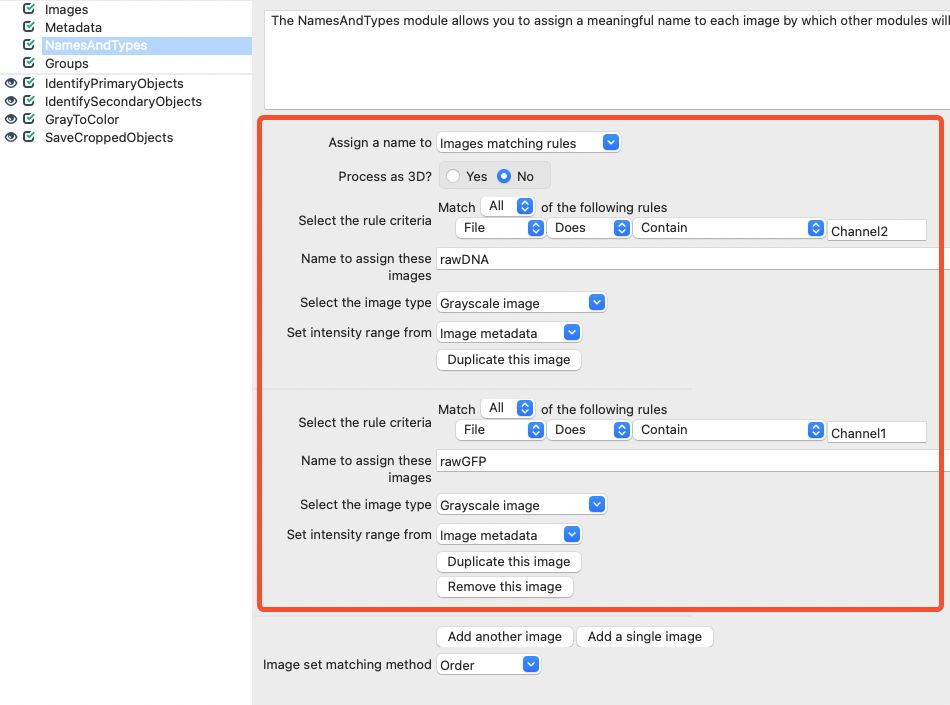
Fig. 20 Within the NamesAndTypes module, assign appropriate names for the DNA and GFP channels.#
2. IdentifyPrimaryObjects#
Then, add a IdentifyPrimaryObjects module and set rawDNA as the input image. Name this primary object Nuclei. Adjust the parameters accordingly so an appropriate segmentation is achieved while using test mode.
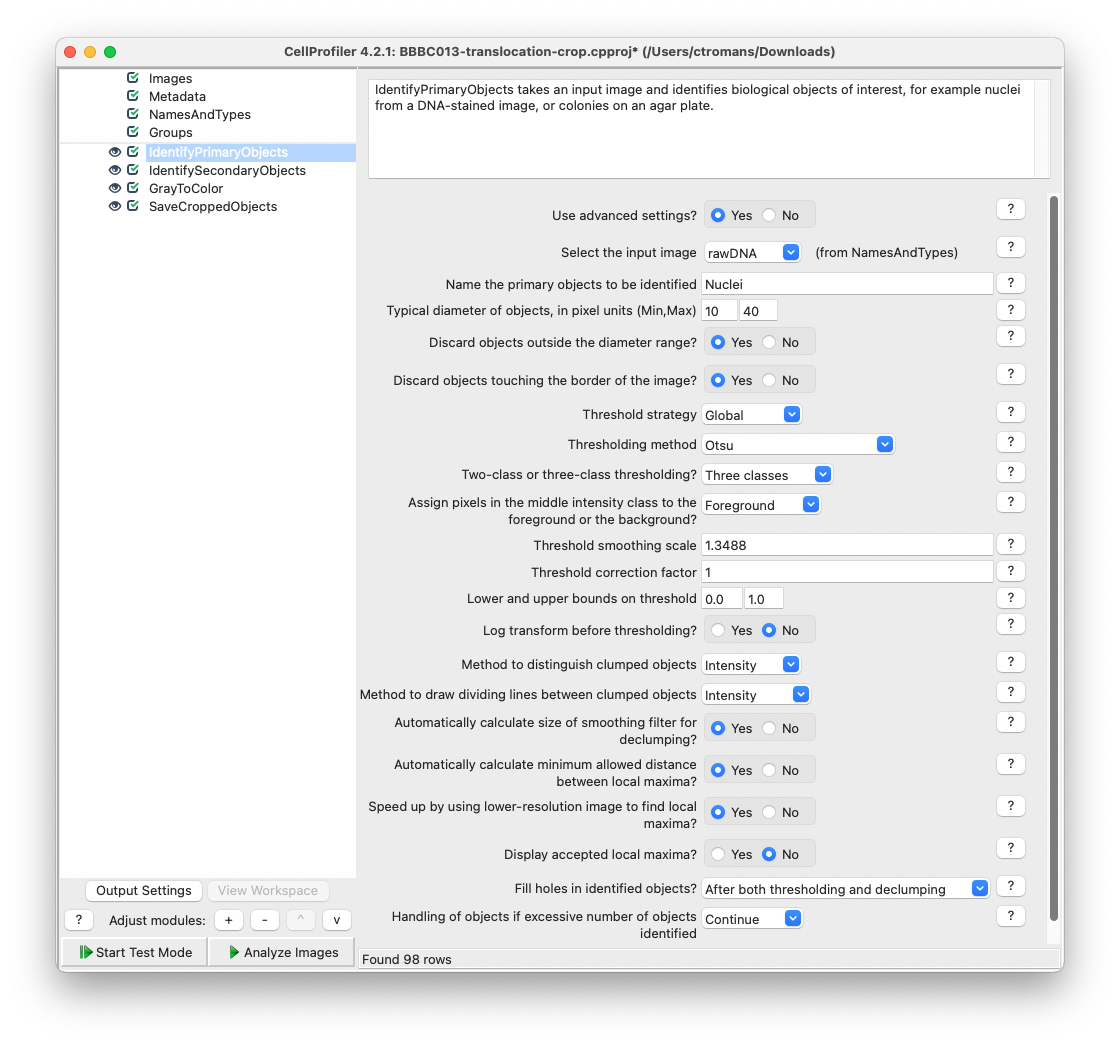
Fig. 21 Add a IdentifyPrimaryObjects module and adjust the parameters to achieve adequate object segmentation of the rawDNA image.#
3. IdentifySecondaryObjects#
Add a IdentifySecondaryObjects module and select rawGFP as the input image and Nuclei as the input objects. Name this Secondary object Cells. Since the cytoplasmic signal is inconsistent between cells and can therefore not be used to determine cytoplasmic segmentation, we will use the Distance - N method to identify secondary objects, with a distance of 10.
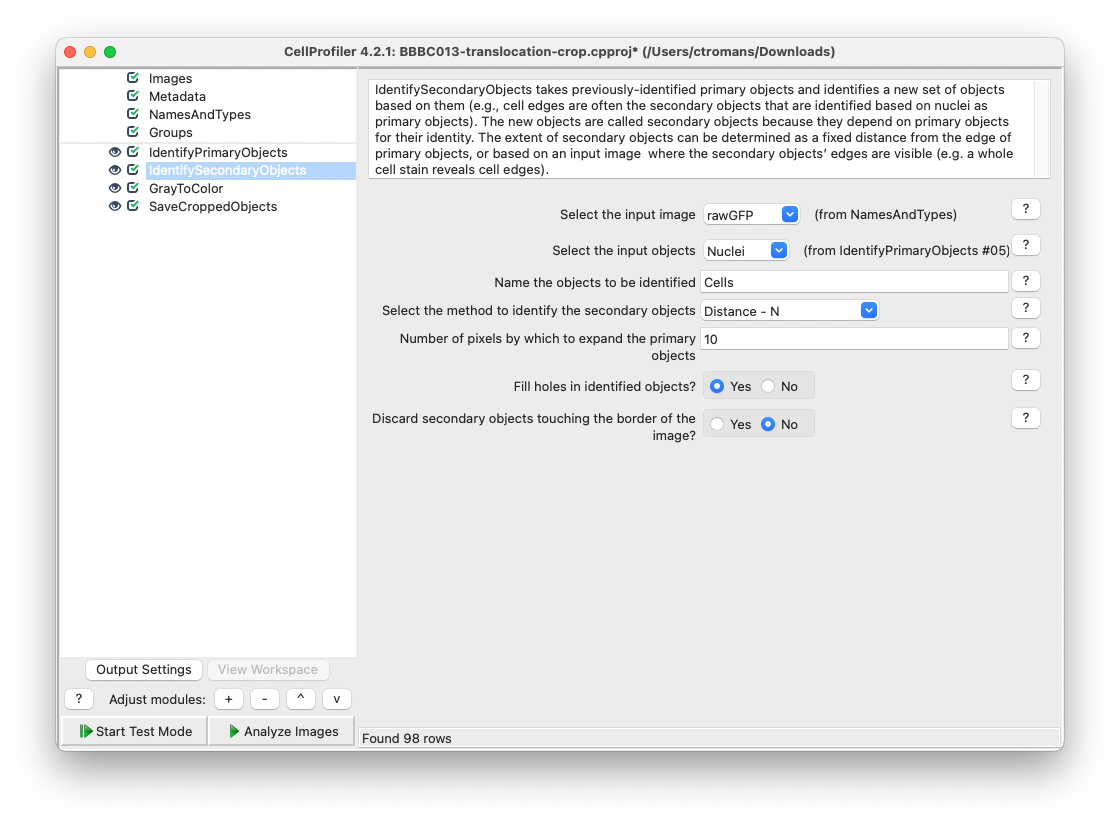
Fig. 22 Add a IdentifySecondaryObjects module using the rawGFP as an input image and Nuclei as in input object.#
4. GrayToColor#
Now, we will create a multichannel RGB image using the input rawDNA and rawGFP images. Add a GrayToColor module and select rawGFP to be colored green and rawDNA to be colored blue. Name the output image GFPandDNA.
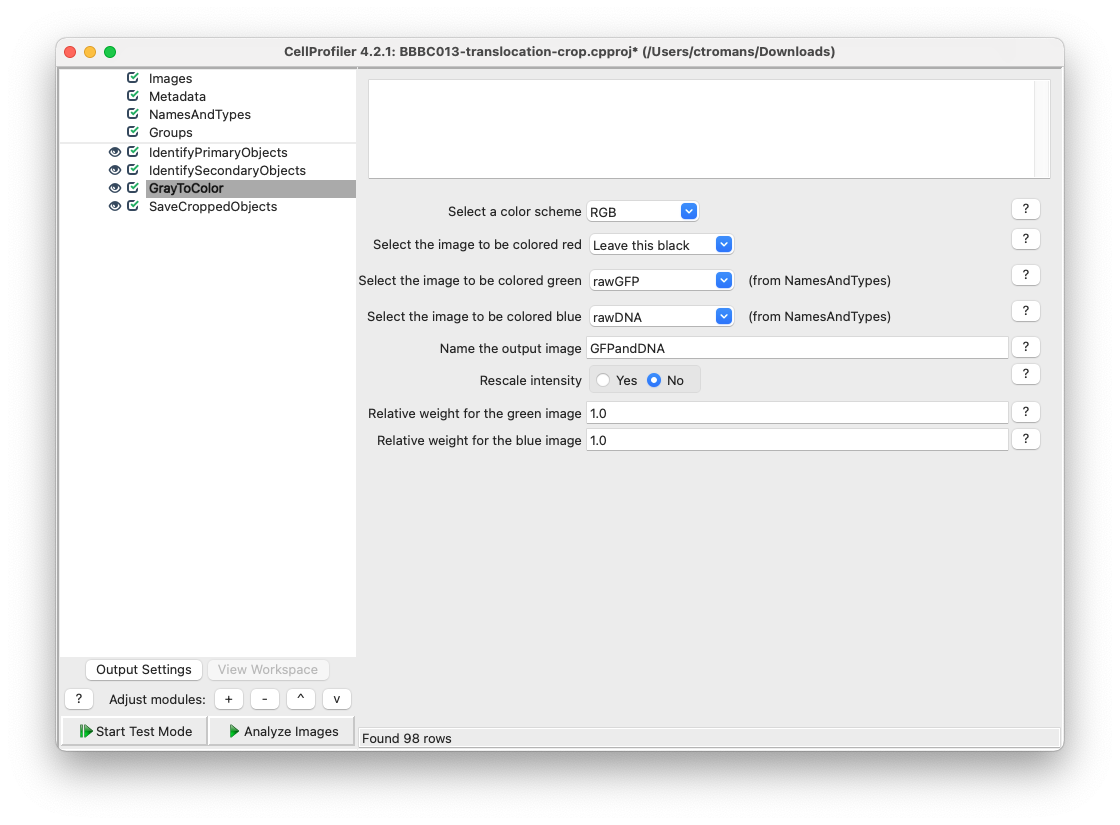
Fig. 23 Add a ColorToGray module and set the rawGFP as green and rawDNA as blue.#
5. SaveCroppedObjects#
Next, add a SaveCroppedObjects module and select the Cells objects from IdentifySecondaryObjects. For the Image to crop setting, select GFPandDNA created by the ColorToGray module. It is this image that will be cropped to individual Cells objects. Select an appropriate directory to save your cropped images.
Note
The below screenshot is from CellProfiler 4.2.2, which includes additional features in the SaveCroppedObjects module. These features allow for crops to be saved with their original input filename and also provides the option to save these crops into nested folders. For versions of CellProfiler earlier than 4.2.2, please see the Extra considerations if using CellProfiler 4.2.1 or earlier section below on how to save cropped objects without these new features.
Extra considerations if using CellProfiler 4.2.1 or earlier
CellProfiler versions 4.2.1 and earlier can also be adjusted to save cropped images into subdirectories named after the input filename. This can be achieved by enabling metadata extraction and using the extracted filename in the subfolder output path in SaveCroppedObjects. Below are the changes to the above mentioned pipeline to achieve this result. The above pipeline that has been modified for CellProfiler 4.2.1 can be downloaded here.
1. Enable metadata extraction
In the Metadata input module, select Yes on the Extract Metadata option. In the field marked Regular expression to extract from file name input the regular expression \-(?P<FileName>.*)\.. This will extract the information after the first - and before the last . from the image filenames in the BBBC013 dataset.
Why extract metadata for this portion of the filename?
For each image there are two individual filenames, representing either channel1 or channel2. If you choose to extract the entire filename, SaveCroppedObjects will be unable to reconcile which filename to use and instead use None. By using the regular expression mentioned above, the extracted filename will be the same across the two channels.
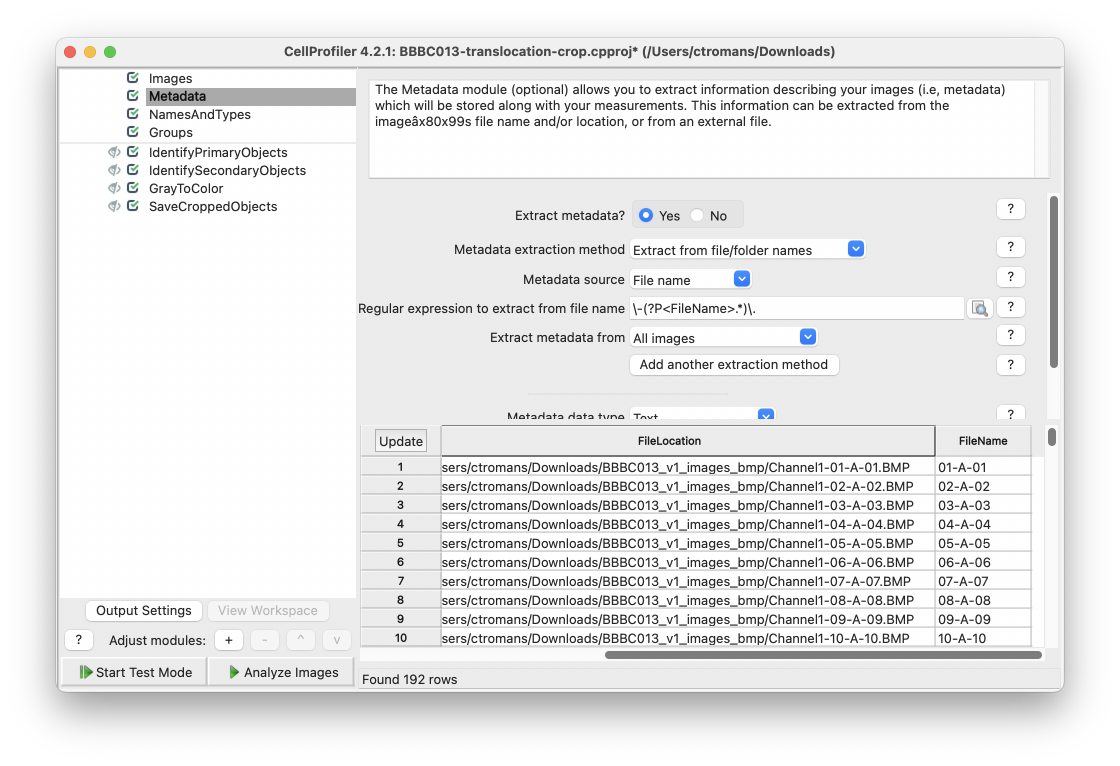
Fig. 24 Use the regular expression \-(?P<FileName>.*)\. to extract an appropriate filename from the BBBC013 images.#
2. SaveCroppedObjects
Within SaveCroppedObjects, select Default Output Folder sub-folder and then right-click in the sub-folder text box and select FileName.
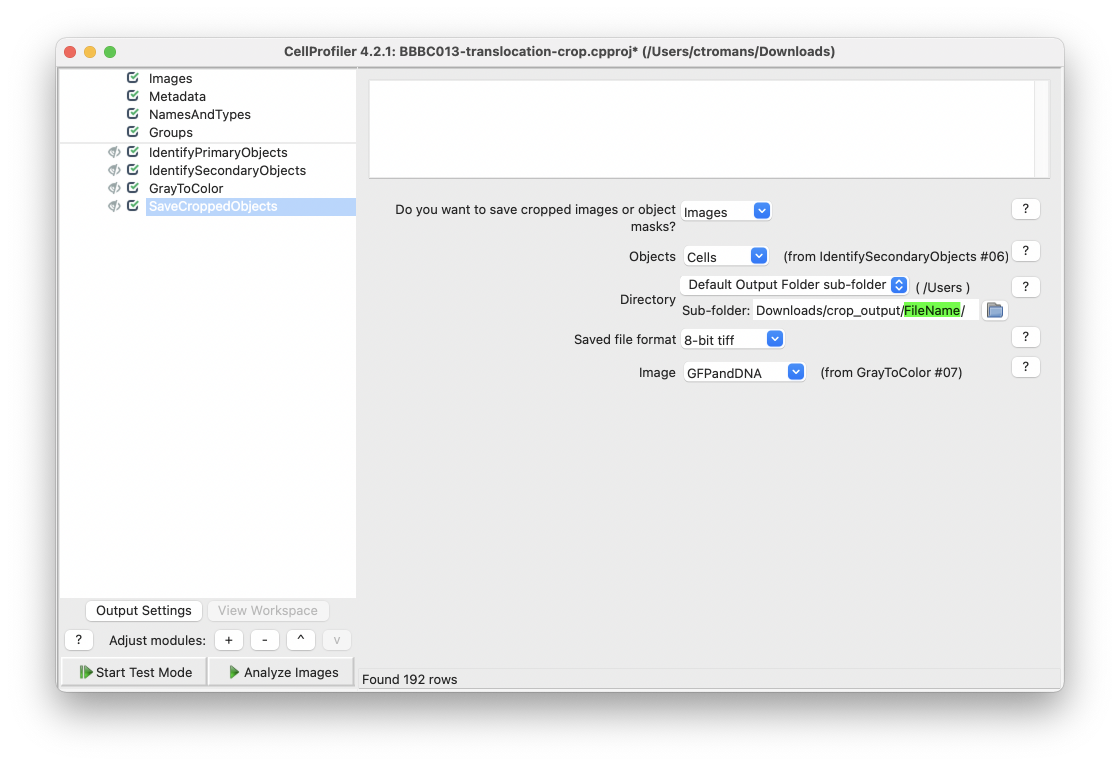
Fig. 25 Right click within the Sub-folder text box and select FileName, as defined in the Metadata module earlier.#
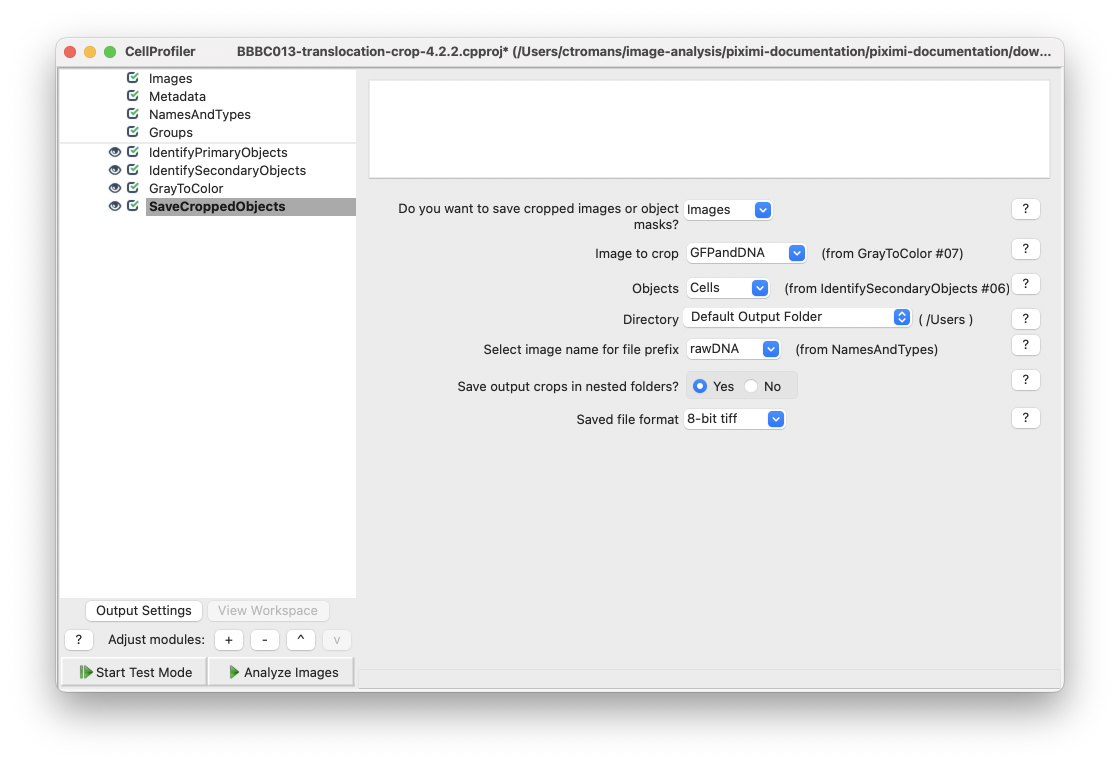
Fig. 26 Add a SaveCroppedObjects module and select your desired Object and the Image to crop. In CellProfiler 4.2.2 you can select to save crops into nested folders with the input image filename.#
6. Next steps#
Now that you have cropped images of your cells, you can follow the image classification guide and adjust it to accommodate your image crops. The crops of the BBBC013 dataset represents a 3-class problem: NuclearGFP, CytoplasmicGFP and NoGFP. When categorizing your images into these distinct categories, explore how categorizing more images into each of these classes can impact training performance.
Copyright
The BBBC013 images used here are licensed under a Creative Commons Attribution 3.0 Unported License by Ilya Ravkin.